|
www.tlab.it
GraphML files
GraphML
format, based on XML
(Extensible Markup Language), allows us to describe the structural
properties of graphs and interchange files between applications
(see http://graphml.graphdrawing.org/).
The T-LAB
function which allows us to export GraphML files
is Sequence Analysis.
Four steps are all we need to do this:
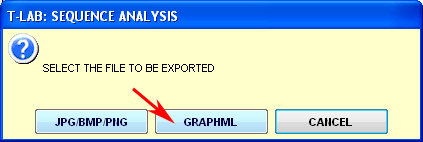
1 - click on
the Save Graphs button of the
T-LAB
function;

2 - select
the graph to be exported (if required);
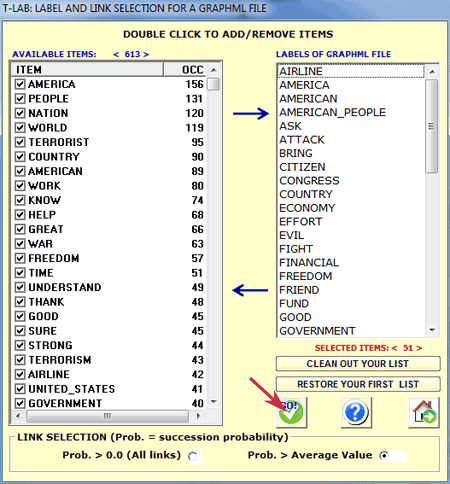
3 - select
the nodes and the edges to be included (if required);
4 - click on
GO to create the GraphML file
(see below).
Two kinds of GraphML
files are exported by T-LAB:
- nomefile.graphml, including only
the node labels and their corresponding
edges;
-
nomefile.EdLab.graphml, which includes the association
measures too.
Both files are ready to be edited and customized by
a GraphML editor like the yEd
software - produced by yWorks -
which is available as a free download (see http://www.yworks.com/en/products_yed_about.htm
).
Follow two examples obtained by editing with
yEd some GraphML
T-LAB
outputs:
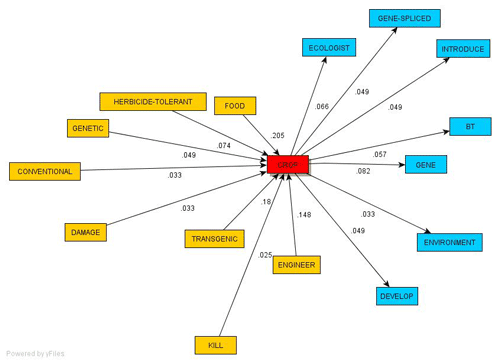
1-
T-LAB
--> Sequence Analysis; yEd:
Layout --> Organic --> Classic
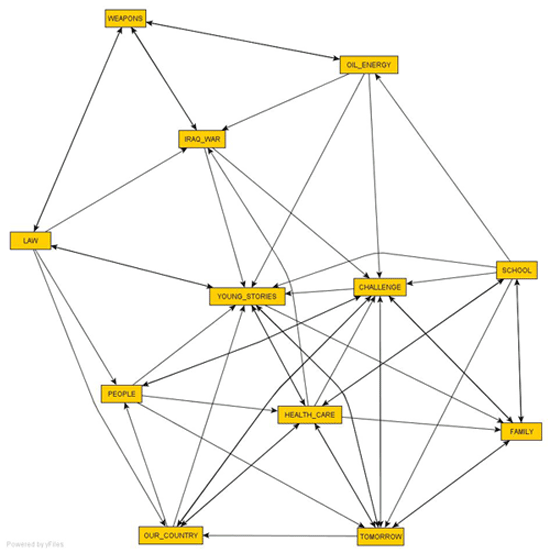
2 -
T-LAB
--> Sequence Analysis; yEd:
Layout --> Organic --> Classic; Edge Router --> Organic
--> Classic
|